Breadcrumb
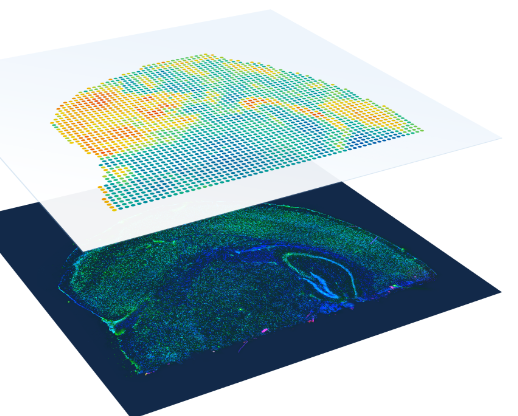
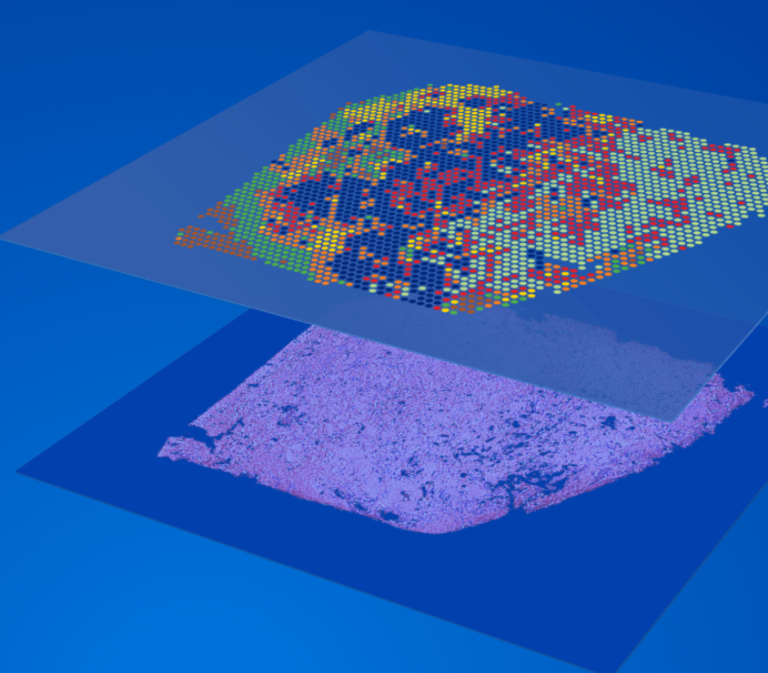
10X Genomics Visium
Each Visium project is unique, the volume, timing, tissue conditions, and research questions of planning varies. The Iowa NeuroBank Core has successfully processed brain, eye, kidney, heart, soft knee tissue, and tail bone across species (human, macaque, mouse, rabbit, and C. elegans) on various Visium platforms for internal and external investigators.
We have been offering 10x Genomics' Visium Spatial Transcriptomics Service and being part of 10X's Visium Enabled Program (VEP) since 2021.
- When all the samples are available, please contact the Core prior to coordinate timing of availability.
Step 1 - Scheduling | Schedule a general discussion by submitting a Visium Consultation Request. |
|---|---|
Step 2 - Prepare your samples (Optional) | Schedule a sample readiness evaluation through the Iowa NeuroBank Core.Histology, custom extraction & purification, whole slide imaging, and expert consultation for the embedded block evaluation, RNA quality assessment, tissue adhesion evaluation, and optimization options. |
Step 3 - Request Quote | Select the services you will use on a Visium Quote Request form. |
Step 4 - Fill out Submission Form | Complete the 10x Genomics Visium Sample Submission Form. |
Step 5 - Drop off your samples | Bring samples to Iowa NeuroBank Core for processing. |
Features:
- Flexible Starting Material: blocks or slides with broad range of RNA quality
- Standard (6.5 mm x 6.5 mm) and larger capture area (11 mm x 11 mm) slides to get more data
- The spatial resolution of Visium is based on the 2-micron squares or 55-micron spot sizes, providing an average resolution of 1 to 10 cells per spot.
- Probe-based Visium platforms: The spatial barcode assigned to the spot is incorporated during cDNA synthesis and enables gene expression data to be mapped back to its location within the tissue. Fastq Data can be processed with the 10X SpaceRanger analysis software, visualized with the Loupe Browser software, and analyzed with the Partek Flow software.
Sample requirements and workflow for Spatial Gene Expression Analysis:
Fresh Frozen Tissue
- Schedule a consultation meeting with us
- Freshly obtained tissue should be snap frozen to prevent RNA degradation.
- The recommended freezing method uses an isopentane and liquid nitrogen bath (equipment available in the Core)
- RNA quality of the tissue block should be assessed prior to sectioning
- Submit your frozen tissue samples for QC (RNA extraction service available in the Core)
- Section your frozen blocks for tissue optimization from each new tissue type for a lab prior to scheduling a Visium GEX appointment.
- Check that the morphology of the 10- µm sections is good enough for cell type annotations. This can be assessed by H&E staining on the SuperFrost Plus Slides.
- Schedule a time to submit your Visium slides with frozen sections
- Staining, Imaging, Permeabilization, cDNA Synthesis, Purification
- Sample and Data Transfer for QC, library construction, sequencing, and data analysis.
FFPE and Fixed Frozen Tissue
- Species Specific – only human and mouse samples currently supported
- Schedule a consultation meeting with us
- Submit your frozen tissue sample for QC (DV200 > 30%; percentage of fragments >200 nucleotides)
- Section your FFPE blocks
- Schedule a time to submit your Visium slide with FFPE sections
- Deparaffinization, Staining, Imaging, Decrosslinking, CytAssist, Re-imaging, Probe Hybridization (18,000 human and 21,000 mouse genes), Ligation, Library Construction Permeabilization, cDNA Synthesis, purification, QC
- Sample and Data Transfer for Sequencing and data analysis.
Questions?
Please email Queena Lin at li-chun-lin@uiow.edu for more information or see the details.